Projects
| Dynamic Networks | RNA-Seq | Clinical Diagnostics | SeqAn |
Dynamic Regulatory Networks
My primary research at Carnegie Mellon University is about the reconstruction of dynamic regulatory networks. Current models of gene regulatory networks are often constructed as a static snapshot of the regulatory wiring in cells. I am working on methods that can dynamically rewire the network connections modeling transcriptional and posttranscriptional factors through the integration of binding data (e.g. Chip-Seq) and gene expression data. In addition, I am trying to utilize
transcript expression level measurements with RNA-Seq to improve the resolution for reconstruction of dynamic regulatory networks.
Dynamic miRNA-TF networks in lung diseases
We are working on the dynamic rewiring of regulatory networks in lung development to predict new regulators in lung diseases. Together with Naftali Kaminskis lab at UPMC we study time series expression data of miRNAs and mRNAs in mouse lungs.
RNA-Seq

I am involved in different projects that improve the analysis of RNA-Seq data. Oases is an accurate de novo transcriptome assembler that uses an explicit alternative splicing model and is able to assemble full length mRNAs from RNA-Seq data without the need of mapping the reads to a reference sequence. The software is freely available and is based on parts of the Velvet genome assembler.
MH Schulz*, DR Zerbino*, M Vingron and E Birney (2012)
Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels (2012)
Bioinformatics 28 (8): 1086-1092 [full text]
The quality of RNA-Seq downstream analysis like de novo transcriptome assembly is diminished by errors introduced during the read sequencing process. We have developed the SEECER software that corrects mismatch and indel errors in non-uniform sequencing data sets, for example RNA-Seq data. If a genomic reference is available as well as transcript annotation, software distributed in the Solas package can be used to infer new alternative splicing events. Also reliable estimates of isoform expression levels for a gene can be computed using the POEM algorithm (see picture above % values on the right).
H Richard*, MH Schulz*, M Sultan*, A Nürnberger, S Schrinner, D Balzereit, E Dagand, A Rasche, H Lehrach, M Vingron, SA Haas, and ML Yaspo (2010)Prediction of alternative isoforms from exon expression levels in RNA-Seq experiments
Nucleic Acids Research, 38 (10):e112 [full text]
*shared first authorship Small RNAs in malaria
Malaria is a serious disease that kills approximately one million people per year worldwide and is caused by different Plasmodium species. Together with the Chakrabarti lab from CMU I am investigating the role of small RNAs in virulence and pathogenesis of Plasmodium falciparum.
Transcriptome analysis of the sea cucumber
The sea cucumber is a remarkable animal that basically does not die. Therefore it is a great model organism to study aging. Together with Veronica Hinmans lab we have produced the first comprehensive transcriptome assembly of sea cucumber development.
Clinical Diagnostics
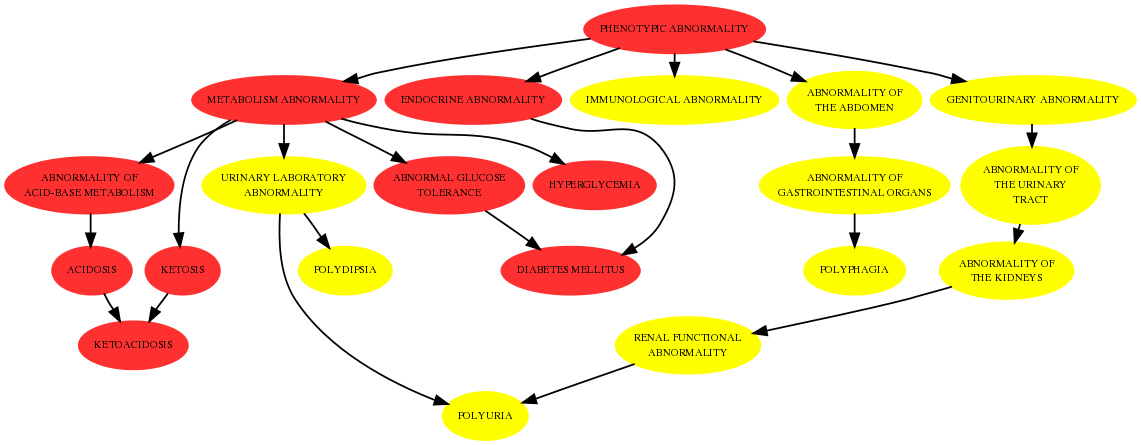
Using the Human Phenotype Ontology (HPO) we develop methods for the diagnosis of diseases using observed phenotypes in patients. We have developed a new procedure to rank potential causal diseases using p-values of semantic similarity measures between terms of the HPO . The most succesful measure can be tested online using the Phenomizer web application. It is even possible to compute the exact p-values with efficient algorithms, which was shown to outperform random sampling presented in BMC Bioinformatics. Extending the applicability of these methods as well as the considerations of annotation errors are directions of our future research.
S Köhler, MH Schulz, P Krawitz, S Bauer, S Dölken,
CE Ott, C Mundlos, D Horn, S Mundlos and
PN Robinson (2009)
Clinical Diagnostics with Semantic Similarity Searches in Ontologies
The American Journal of Human Genetics, 85 (4):457-64 [full text]
MH Schulz, S Köhler, S Bauer and PN Robinson
Exact score distribution computation for ontological similarity searches
BMC Bioinformatics, 12:441 [full text]
SeqAn - C++ Library for Sequence Analysis

Bioinformatic software is under the permanent need to adapt to the increasing throughput of modern technologies, especially Next-Generation Sequencing. It is therefore essential that open source libraries exist that supply the researcher with up-to-date implementations for common tasks in Sequence Analysis. To often researchers resort to ad-hoc implementations and non-optimized algorithms due to lack of availability and time. SeqAn is a C++ template library for Biological Sequence Analysis that has been growing considerably over the last years and contains efficient implementations of all major building blocks for Sequence Analysis. I contribute to SeqAn to improve it further. Thus far my most important contributions are data mining algorithms and algorithms for construction of variable order Markov chains.
MH Schulz, D Weese, T Rausch, A Döring, K Reinert and M VingronFast and adaptive variable order Markov chain construction
Proceedings WABI 2008, Springer LNCS, Volume 5251 [Full text]
D Weese and MH Schulz
Efficient string mining under constraints via the deferred frequency index
Industrial Conference for Data Mining (ICDM 2008), LNAI 5077, pp. 374-388
[Full text]