Back to "Homepage of Judith Klein-Seetharaman"
Back to "G-protein coupled receptors"
G-protein coupled receptor reviews
- review on class A GPCR, i.e. rhodopsin
- comparison of rhodopsin structure with previous computational models for TM domain (~3.2Å rmsd for ~200 alpha-carbons)
- review of helix movements: activation model

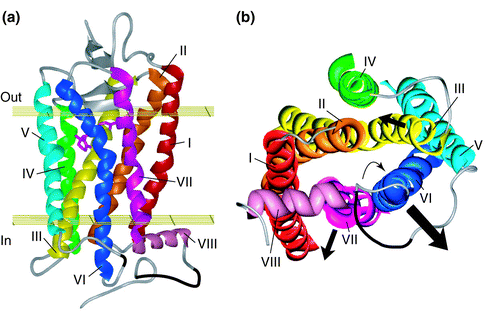
Figure: Helix movement upon activation of rhodopsin. Figure is Figure 1 in Meng and Bourne (2001) Trends in Pharmacological Sciences: Receptor activation: what does the rhodopsin structure tell us
- review of crosslinking of the chromophore
- review of disulfide bond crosslinking data in rhodopsin (Table 1) and their effect on Gt-activation
- review of EPR results
Visiers, I., Ballesteros, J.A. and Weinstein, H. (2002) Methods in Enzymology 343, 329-371.
- review on 5HT2A, rhodopsin, opioid receptors
- review of "aromatic cluster" and "arginine cage" for activation mechanism
- comparison of rhodopsin crystal structure with other Class A GPCR, i.e. amine GPCR, i.e. dopamine D2 receptor
- structural mimicry in GPCR: despite different amino acid sequence, and different "microdomains", several structural features conserved:
- predictions for dopamine (or amine in general) receptor that are well compatible with rhodopsin structure:
1. ends of helices, i.e. CP ends cannot be predicted by hydrophobic plot or moment, because Arg and Lys contenrate tat the CP boundary of membrane proteins where they interact with the lipid at th ephospolipid headgroups - thus, although the precise position of positive aa is not conserved, the fact that there are positive charges at the CP ends is conserved (in amine GPCR, there are 3times more positive than negative amino acids at CP ends of helices), thus the position of positive amino acids can be used to predict the ends of the helices
2. surface of binding-site crevice by cysteine-accessibility method
3. deviations from regular helical structure
4. lipid exposure of residues (predictions based on physicochemical properties plus experiments)
5. Tertiary structure interactions in amine receptors compatible with rhodopsin: cysteine crosslinking, double revertant mutations, Zinc-his binding, spin labeling , ligand related, FTIR
1. disulfide crosslinking in muscarinic receptor ACM2 and ACM3 (Zeng, Hopp et al. 1999) and zinc his binding ACM1 Lu and Hulme 2000
3.54-6.29 (ACM2), 3.54-6.30...6.36 (ACM3), 3.43-6.44,7.49 (ACM1), 6.44-7.49
A. disulfide paper:
method: visualization of receptor by antibody tag in cell lysates
added Factor Xa cleavage site in third CP loop, just before the second cysteines, so that disulfide crosslinked receptor migrates at same position as native, while non-crosslinked migrates at fragment positions
analogous to the Farrens study - one cysteine at CP end of helix 3, and set of second cysteine position at CP end of helix 6
8 out of 10 mutants formed disulfide bond with copper-phenantroline
functional consequence: blocks activation drastically where disulfides formed (same as in rhodopsin)
notion of dynamics in CP face that allows disulfide bonds to form
B. zinc his binding paper:
two residues in TM3 of M1 mAChR cause constitutive activation when mutated: Leu116 and Ser120
authors investigated what residues are in contact to these by Zn approach
the residues are in helices 3, 5,6,7 all facing the inside of the bundle (positions in middle of helices, somewhat towards the CP side)
the effects on signal transduction are not very clear
2. zinc-his binding in NK1 tachykinin receptor (NK1R) Elling and Schwartz, 1996; Elling et al., 1995
based on zinc binding inactivating the binding of agonist "antagonistic zinc binding"
2.64-3.28; 3.29-5.35; 5.35-6.59, 5.39-6.55,6.56,6.59
3. kappa-opioid receptor Thirstrup et al., 1996
30% identity to NK1R
engineered zinc binding sites ananlogous to those in NK1R, at bottom of helices 5 and 6, gave analogous results (ligand binding not affected, but new Zn binding site created)
conclusion: overall architecture same
4. zinc his binding in B2AR Elling et al., 1999; and Scheikh et al., 1999
3.32-7.39 (Elling et al)
Elling et al. engineered Zn-chelator complex binding sites that ACTIVATE the receptor: signal transduction is not induced by ligand binding, but by addition of metal-chelator complexes: distance constraint on the ACTIVE conformation!! Distance from Zn to coordinating atoms is 2 A, cna therefor be accomodated by the two residues (Cbetas were 8 A apart in Baldwin and Hubbard models)
unclear if ligands bring helices closer together or in fact keep them further apart than in the inactive state
metal binding as potential drug targets that could be used to evaluate
1. what physiological function an orphan receptor has
2. if a particular GPCR is a good drug target
sites at amine-binding Asp113 in TM3 and Asn312 in TM7 (EC side)

Figure taken from Elling
et al., 1999: View from EC side. Molecular models of rhodopsin (Upper)
and the [His-113,Cys-312] beta2-adrenergic receptor (Lower). The C-alphas
of residues III:08 and VII:06 (constituting the activating metal-ion site in the
beta2-AR) are indicated by large green spheres. In the [His-113,Cys-312]
beta2-AR His-113 (HisIII:08) and Cys-312 (VII:06) are indicated in stick models
and the proposed coordination of a metal-ion between these two residues and the
two nitrogens of 1,10-phenanthroline is also indicated. The Calphas of residues
III:04 and VII:10 (the two main residues involved in retinal binding) are
indicated by large red spheres.
3.53-6.30,6.31,6.34 (Sheikh et al.)
done in cell membrane preparations, G protein activation assay: dependence on Zn concentration taken as indication for disulfide bond effect
-> shows movement of helices 3 and 6 required for activation, because with Zn G protein activation is blocked.
5. Zn-His in PTHR (parathyroid hormone receptor) Sheikh et al., 1999
analogous to the study of B2AR by Sheikh et al., 1999 above, although PTHR and B2AR are from different GPCR classes (no sequence homology)!!
3.53-6.30,6.31,6.34 (Sheikh et al.)
done in cell membrane preparations, G protein activation assay: dependence on Zn concentration taken as indication for disulfide bond effect
-> shows movement of helices 3 and 6 required for activation, because with Zn G protein activation is blocked.
6. Disulfides in NK1R Elling, C. E., U. Raffetseder, et al. (2000). "Disulfide bridge engineering in the tachykinin NK1 receptor." Biochemistry 39(4): 667-75.
in extracellular domain: Nt-EC3 and at EC ends of TM5-TM6
disulfide bond formation monitored by loss of agonist binding, and recovery of binding upon DTT addition
disulfide bonds did not form as much at ends of TM5-TM6 as observed previously with Zn-binding sites
- predictions for dopamine (or amine in general) receptor that differ from rhodopsin crystal structure:
1. helix 4
- "The extracellular and cytoplasmic domains of GPCRs are widely divergent, thus precluding any direct comparison between rhodopsin and other GPCRs". Therefore focus on TM domains.
- Second extracellular loop highly variable in classA family, but from 4.59-4.68 very conserved among functionally related receptors, e.g. in alpha2-adrenergic receptor, ligand crosslinks here, loop thought to contact ligand in other receptors of family A as well, e.g. specificity of particular adrenergic ligands is determined here (can be changed by site directed mutagenesis)
Generality of Tryptophans in GPCR family:
- highly conserved Trp4.50 (trp161 in rhodopsin) faces lipid in rhodopsin, but the binding site crevice in dopamine receptor
- Trp6.48 (Trp265 in rhodopsin) is part of a cluster of aromatic residues n TM5 and TM6 that interact with the aromatic ring of ligands in the beta2-adrenergic receptor
-Trp6.48 in 5H1B, 5H2A receptors found in SASA calculations to be protected by retinal and implicated in ligand binding
- review of extracellular loop 2: beta-strand (186-189) runs parallel along the retinal chain, strand stabilized by disulfide bond, provides contact with retinal through Glu181 and Tyr191, function as "cap or lid which traps the 11-cis retinal in its binding pocket" or "to regulate the stability of the active state
- some review of activation
- some review of differences between rhodopsin and bacteriorhodopsins
- Classification of GPCR is uncertain
1. GPCRs do not segregate into groups based on the G protein to which they couple, and many receptor can signal via G-protein independent mechanisms
2. GPCR are very divergent in sequence, even within one organism
- Length distribution of N-terminus: 2 to 2500
- "The only possible structural element that might be conserved among all 7TM is a disulfide bond"
- crystal structures of N-terminal domains
1. glutamate receptor: Kunishima, N., Y. Shimada, et al. (2000). "Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor." Nature 407(6807): 971-7.
2. mouse Frizzled 8 protein which binds Wnt: Dann, C. E., J. C. Hsieh, et al. (2001). "Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains." Nature 412(6842): 86-90. (get ref)
- detailed discussion of the residues important for activation of rhodopsin
- network of hydrogen bonds in the middle of the bundle, constraining rhodopsin in the ground state
Asn1.50-Asp2.50; Trp4.50 (highly conserved in GPCR)
"for GPCR that do not have these residues, a different set of interhelical constraints around these regions is modified by ligand binding during activation"
- other constraints:
at CP ends of helices, bundle constrained mainly by hydrophobic interactions, arranged in 2 layers stacked perpendicular to the helical axes
Layer 1. above network of H-bonds (see above)
hydrophobic interactions on the CP side of helices 7, 3 and 2 contain L2.46 (highly conserved) need to break during activation (see also constitutively active mutants in this area of receptors)
Layer 2. surrounds E/D R Y (3.49-3.51) motif
R3.50 is most conserved, H-bond to E/D.349 and interacts with polar sidechains on the CP side of helix 6
Y3.51 exposed to outside, interacts with Pro142
=> layers of hydrophobic interactions must rearrange during activation
Summary: "The constrains on the heptahelical bundle of rhodopsin can be described as the sum of electrostatic and hydrophobic interactions tacked along the CP surface of rhodopsin"
- chromophore trigger:
- neutralization of salt bridge between PSB and counter ion Glu113(3.28) [similar ionic interaction between cationic group of epinephrine and acidic sidechain 3.32 in adreneregic receptors]
- effect of charge neutralization: helix 7 is not stabilized anymore
salt bridge and other interhelical constraints stabilize NPXXY (7.49-7.53) motif bend in helix 7, this is broken
set of hydrogen bonds among helix 7 (peptide CO of A7.46, N1.50 and D2.50) are broken
- comparison to bacteriorhodopsin:
- Role of Trp265:
analogy: rearrangment in bR is triggered initially by steric conflict between C13 methyl group of retinal and the Trp182 in helix 6 Edman, K., P. Nollert, et al. (1999). "High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle." Nature 401(6755): 822-6.
here: C13 methyl in rhodopsin interacts with Trp265 in ground state, indole ring rotates during photoactivation Lin, S. W. and T. P. Sakmar (1996). "Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state." Biochemistry 35(34): 11149-59.
- large retinal motion will trigger change in constraints among helices 2,3,4 and 5 including His211 and Glu122
Gether, U. (2000). "Uncovering molecular mechanisms involved in activation of G protein-coupled receptors." Endocr Rev 21(1): 90-113.
- supercedes older review by Gether and Kobilka (1998), see below. This one is much more detailed. Excellent Review!
- paper speculates on dynamics aspects of activation mechanism
- in February 2000, there were more than 1000 sequences [how many sequences are there now? How many odorant sequences are there? From how many different organisms do we have sequences and how does the distribution look like along organisms?]
- the largest superfamily of proteins in the body (i.e. the human genome)
- complexity of GPCR signaling - not only via heterotrimeric G-proteins (see GPCR protein protein interactions)
- crosstalk to other signaling cascades (see GPCR protein protein interactions)
- review of ligand binding domain (see GPCR ligand binding)
- review of activation mechanism (see GPCR activation)
- review of receptor dimerization (see GPCR dimer)
- overview of properties and conserved residues of Class A, B, C - very nice figure: Figure 1
PLACE FIGURE 1 HERE!
- "experimental studies aimed at probing tertiary structure relations in GPCRs" including "identification of several distance constraints in the receptor structure":
1. chimera alpha2/beta2 adrenergic receptors and muscarinic M2/M5 receptors used for identification of helix I&VII close proximity, reviewed in Gether (2000):
Suryanarayana, S., M. von Zastrow, and B.K. Kobilka, Identification of intramolecular interactions in adrenergic receptors. J Biol Chem, 1992. 267(31): p. 21991-4.
Pittel, Z. and J. Wess, Intramolecular interactions in muscarinic acetylcholine receptors studied with chimeric m2/m5 receptors. Mol Pharmacol, 1994. 45(1): p. 61-4.
Liu, J., T. Schoneberg, M. van Rhee, and J. Wess, Mutational analysis of the relative orientation of transmembrane helices I and VII in G protein-coupled receptors. J Biol Chem, 1995. 270(33): p. 19532-9.
Mizobe, T., M. Maze, V. Lam, S. Suryanarayana, and B.K. Kobilka, Arrangement of transmembrane domains in adrenergic receptors. Similarity to bacteriorhodopsin. J Biol Chem, 1996. 271(5): p. 2387-9.
2. histidine zinc(II( binding sites in neurokinin 1 (NK1) (substance P) and kappa-opioid receptors used for relative orientation of helix III relative to II and V, reviewed in Gether (2000):
Elling, C.E. and T.W. Schwartz, Connectivity and orientation of the seven helical bundle in the tachykinin NK-1 receptor probed by zinc site engineering. Embo J, 1996. 15(22): p. 6213-9.
Elling, C.E., S.M. Nielsen, and T.W. Schwartz, Conversion of antagonist-binding site to metal-ion site in the tachykinin NK-1 receptor. Nature, 1995. 374(6517): p. 74-7.
Thirstrup, K., C.E. Elling, S.A. Hjorth, and T.W. Schwartz, Construction of a high affinity zinc switch in the kappa-opioid receptor. J Biol Chem, 1996. 271(14): p. 7875-8.
3. Disulfide bonds in rhodopsin
all our references
Yu, H., M. Kono, T.D. McKee, and D.D. Oprian, A general method for mapping tertiary contacts between amino acid residues in membrane-embedded proteins. Biochemistry, 1995. 34(46): p. 14963-9.
4. Disulfide bonds in M3 muscarinic receptor
Zeng, F.Y., A. Hopp, A. Soldner, and J. Wess, Use of a disulfide cross-linking strategy to study muscarinic receptor structure and mechanisms of activation. J Biol Chem, 1999. 274(23): p. 16629-40.
5. Fluorescence energy transfer measurement of distances between fluorescent peptide antagonist and a fluorescent amino acid in tachykinin NK2 receptor
Turcatti, G., K. Nemeth, M.D. Edgerton, U. Meseth, F. Talabot, M. Peitsch, J. Knowles, H. Vogel, and A. Chollet, Probing the structure and function of the tachykinin neurokinin-2 receptor through biosynthetic incorporation of fluorescent amino acids at specific sites. J Biol Chem, 1996. 271(33): p. 19991-8.
- tertiary structure by cysteine accessibility method (in other GPCR means mostly loss of ligand binding upon cysteine derivatization)
1. rhodopsin (different purpose than the other studies below)
all our papers
2. Dopamine D2 receptor (helices II, III, V, VI and VII) --> using charged
methanethiosulfonate (MTS) identified binding site and Proline kinks in helices
(PVI.15/6.50 and PVII.17/7.50)
Fu, D., J.A. Ballesteros, H. Weinstein, J. Chen, and J.A. Javitch, Residues in the seventh membrane-spanning segment of the dopamine D2 receptor accessible in the binding-site crevice. Biochemistry, 1996. 35(35): p. 11278-85.
Javitch, J.A., D. Fu, and J. Chen, Residues in the fifth membrane-spanning segment of the dopamine D2 receptor exposed in the binding-site crevice. Biochemistry, 1995. 34(50): p. 16433-9.
Javitch, J.A., D. Fu, J. Chen, and A. Karlin, Mapping the binding-site crevice of the dopamine D2 receptor by the substituted-cysteine accessibility method. Neuron, 1995. 14(4): p. 825-31.Javitch, J.A., J.A. Ballesteros, H. Weinstein, and J. Chen, A cluster of aromatic residues in the sixth membrane-spanning segment of the dopamine D2 receptor is accessible in the binding-site crevice. Biochemistry, 1998. 37(4): p. 998-1006.
- is superceded by new review by Gether. That one is much more detailed.
- classification of GPCR in 3 families "based on certain key sequences"
- "extended ternary complex model":
ternary complex model = cooperative interactions between GPCR, G protein and agonist
extended = include activation of G protein in the absence of agonists (constitutive activation)
also accounts for the effects of full agonists, partial agonists, natural antagonists and inverse agonists
in the extended model, the receptor exists in an equilibrium of the inactive and the activate state and the efficacy of ligands is a reflection of their ability to alter the equilibrium between these states
model may need to be further extended to account for more complicated relationships
activation of rhodopsin is unique among GPCR because ligand binding is not part of activation process
- "compelling evidence from several different receptors" that there is a stabilizing network of intramolecular interactions constraining the receptor in the inactive conformation (see references 14, 33-36 in Gether and Kobilka review):
1. rhodopsin salt bridge between TM3 and TM7 (disruption causes constitutive activity)
2. follicle-stimulating hormone receptor and luteinizing hormone receptor: interactions between TM5 and TM6 (get ref. Kudo, M., Y. Osuga, et al. (1996). "Transmembrane regions V and VI of the human luteinizing hormone receptor are required for constitutive activation by a mutation in the third intracellular loop." J Biol Chem 271(37): 22470-8.)
3. beta2-adrenergic receptor: TM6 relative to TM3 and TM5 Javitch, J. A., D. Fu, et al. (1997). "Constitutive activation of the beta2 adrenergic receptor alters the orientation of its sixth membrane-spanning segment." J Biol Chem 272(30): 18546-9.
4. NK-1 (substance P) receptor: zinc his links and disulfide crosslinks:
Elling, C. E., S. M. Nielsen, et al. (1995). "Conversion of antagonist-binding site to metal-ion site in the tachykinin NK-1 receptor." Nature 374(6517): 74-7.
Elling, C. E., U. Raffetseder, et al. (2000). "Disulfide bridge engineering in the tachykinin NK1 receptor." Biochemistry 39(4): 667-75.
Elling, C. E., K. Thirstrup, et al. (1997). "Engineering of metal-ion sites as distance constraints in structural and functional analysis of 7TM receptors." Fold Des 2(4): S76-80.
Elling, C. E., K. Thirstrup, et al. (1997). "Metal-ion sites as structural and functional probes of helix-helix interactions in 7TM receptors." Ann N Y Acad Sci 814: 142-51.
Bourne (1997) How receptors talk to trimeric G proteins. Curr. Opin. Cell Biol. 9, 134-142.
- reviews possible mechanisms by which G-proteins bind to receptors
Wilson, S., D.J. Bergsma, J.K. Chambers, A.I. Muir, K.G. Fantom, C. Ellis, P.R. Murdock, N.C. Herrity, and J.M. Stadel, Orphan G-protein-coupled receptors: the next generation of drug targets? Br J Pharmacol, 1998. 125(7): p. 1387-92
- useful review of strategies to classify putative GPCR sequences pharmacologically
- is what they do at SmithKline Beecham
- Pharmacological relevance of GPCR:
1.- table of drugs targeting GPCR
2.- diseases assocated with GPCR mutations:
rhodopsin RP
thyroid stimulating hormone hyperfunctioning thyroid adenomas
lutenizing hormone precious puberty
vasopressin V2 receptor X-inked nephrogenic diabetes
calcum sending hyuperparathyroidism, hypocalciuria, hpercalcemia
parathyroid hormone short limbed dwarfism
beta3-adrenorectpor obesity, NIDDM
growth hormone releasing hormone dwarfism
adrenocorticotropin glucocorticoid deficiency
glucagon diabetes, hypertension
- in most cases the sequence homology of newly identified GPCR sequences is too low to assign them to a particular receptor subfamily
- therefore two other approaches used:
A. reverse molecular pharmacological methodology [Libert, F., G. Vassart, and M. Parmentier, Current developments in G-protein-coupled receptors. Curr Opin Cell Biol, 1991. 3(2): p. 218-23]:
1. identify orphan GPCR by bioinformatic analysis of expressed sequence tag (EST) databases generated by mass random sequencing of cDNA libraries (EST approach is a highly productive route for identifying novel genes, see Adams, M.D., M. Dubnick, A.R. Kerlavage, R. Moreno, J.M. Kelley, T.R. Utterback, J.W. Nagle, C. Fields, and J.C. Venter, Sequence identification of 2,375 human brain genes. Nature, 1992. 355(6361): p. 632-4]
2. obtain full-length cDNA
3. cloning and expression of orphan GPCR in mammalian cells (CHO or HEK293 mostly, but also Xenopus oocytes, melanophores and engineered yeast systems) containing an epitope tag to assess protein expression via FACS or Western blots, e.g. FLAG and HA tags for which antibodies are commercially available)
4. activity measurement by changes in intracellular cAMP or Ca2+ levels using standard methods or via the use of reporter gene assays or better:
5. use a single signal transduction system for many GPCR by co-expression of the receptor wtih the promiscuous G protein Galpha15/16 or with chimeric Gq proteins such as Gqi5 [Conklin, B.R., P. Herzmark, S. Ishida, T.A. Voyno-Yasenetskaya, Y. Sun, Z. Farfel, and H.R. Bourne, Carboxyl-terminal mutations of Gq alpha and Gs alpha that alter the fidelity of receptor activation. Mol Pharmacol, 1996. 50(4): p. 885-90.; Offermanns, S. and M.I. Simon, G alpha 15 and G alpha 16 couple a wide variety of receptors to phospholipase C. J Biol Chem, 1995. 270(25): p. 15175-80]
6. Yeast system: can genetically delete the endogenous GPCR to general null background, then commandeering the endogenous yeast GPCR transduction system, the pheromone mating pathway, to allow coupling of transfected human GPCR and humanized G-proteins to the endogenous signaling machinery (yeast response to growth arrest or to reporter gene expression): Broach, J.R. and J. Thorner, High-throughput screening for drug discovery. Nature, 1996. 384(6604 Suppl): p. 14-6
7. screening cells for function response to cognate or surrogate agonists present in biological extract preparations, peptide libraries and complex compound collections
B. functional genomic methodology:
1. use humanized yeast cells where the yeast GPCR transduction system is engineered to permit function expression and coupling of human GPCR to the endogenous signaling machinery
- in vertebrates, 1000-2000 members >1% of genome including >1000 coding for odorant and pheromone receptors
- in C. elegans 1100 GPCR (5% of genome)
- GPCR among the oldest signal transduction systems since they are present in plants [Plakidou-Dymock, S., D. Dymock, and R. Hooley, A higher plant seven-transmembrane receptor that influences sensitivity to cytokinins. Curr Biol, 1998. 8(6): p. 315-24], yeast [Dohlman, H.G., J. Thorner, M.G. Caron, and R.J. Lefkowitz, Model systems for the study of seven-transmembrane-segment receptors. Annu Rev Biochem, 1991. 60: p. 653-88] and slime mold Dictiostelium [Devreotes, P.N., G protein-linked signaling pathways control the developmental program of Dictyostelium. Neuron, 1994. 12(2): p. 235-41] and protozoa and the earliest diploblastic metazoa [Vernier, P., B. Cardinaud, O. Valdenaire, H. Philippe, and J.D. Vincent, An evolutionary view of drug-receptor interaction: the bioamine receptor family. Trends Pharmacol Sci, 1995. 16(11): p. 375-81; New, D.C. and J.T. Wong, The evidence for G-protein-coupled receptors and heterotrimeric G proteins in protozoa and ancestral metazoa. Biol Signals Recept, 1998. 7(2): p. 98-108]
- review focusing on homo- or hetero-dimers of GPCR possibly via coiled-coil interaction of TM VI, but also with proteins with only 1 TM domain, e.g. nina-A, odr-4 and RAMP
- the 1TM proteins are involved in targeting, function and pharmacology
- GPCR unfaithful to G proteins and interact directly with C-terminus with PDZ containing proteins and Enabled-VASP homology (EVH) like domains:
-beta2-adrenergic receptor contacts via its C-terminal DSLL the first PDZ domain of the Na+/H+ exchanger regulatory factor which plays a role in the regulation of Na+/H+ exchange Hall, R.A., R.T. Premont, C.W. Chow, J.T. Blitzer, J.A. Pitcher, A. Claing, R.H. Stoffel, L.S. Barak, S. Shenolikar, E.J. Weinman, S. Grinstein, and R.J. Lefkowitz, The beta2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H+ exchange. Nature, 1998. 392(6676): p. 626-30
- MUPP1 is a multi PDZ domain protein with unknown function which interacts with 5HT2C [Ullmer, C., K. Schmuck, A. Figge, and H. Lubbert, Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett, 1998. 424(1-2): p. 63-8]
- Homer/vesl are family with an Enabled/VASP homology (EVH)-like domain which interacts with group 1 mGluR [Brakeman, P.R., A.A. Lanahan, R. O'Brien, K. Roche, C.A. Barnes, R.L. Huganir, and P.F. Worley, Homer: a protein that selectively binds metabotropic glutamate receptors. Nature, 1997. 386(6622): p. 284-8]
- Homer1a is upregulated in seizures and in long0erm potentiation, a form of synaptic plasticity which may be involved in memory and seiuzres
- a novel homer ligand PPXXFR in C-termini of mGluR1a and mGluR5 tightly interacts with the EVH domain of all Homer isoforms [Tu, J.C., B. Xiao, J.P. Yuan, A.A. Lanahan, K. Leoffert, M. Li, D.J. Linden, and P.F. Worley, Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron, 1998. 21(4): p. 717-26]
- the homer ligand is also present in the IP2 receptor and ryanodine receptor. Thus it is likely that complexes containing mGluR1a, mGluR3, HOmer1bc, HOmer2,3 and IP2 and possibly ryanodine receptors can be formed
- homer 1a which does not multimerize because of the absence of coiled coil domain that normally homer proteins have allowing them to multimerize, has been shown to disrupt the formation of multivalent complexes between other Omer proteins and mGluR. Thiseffect results in teh inhibition of mGluR induced Ca2+ release from IP2 receptors in Purkinje cells Xiao, B., J.C. Tu, R.S. Petralia, J.P. Yuan, A. Doan, C.D. Breder, A. Ruggiero, A.A. Lanahan, R.J. Wenthold, and P.F. Worley, Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron, 1998. 21(4): p. 707-16.]
- homer proteins are localized in postsynaptic density of synaptic structures, providing evidence that Homer 1a, which is rapidly expressed ruing thigh synaptic activity, may feed back to reduce synaptic function [Tu, J.C., B. Xiao, J.P. Yuan, A.A. Lanahan, K. Leoffert, M. Li, D.J. Linden, and P.F. Worley, Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron, 1998. 21(4): p. 717-26]
Specialized reviews:
1. Thyrotropin Receptor and Disease: Paschke and Ludgate (1997) New England J. of Medicine 337, 1675-1681. Includes some mutations causing constitutive activity.
2. Beta3-adrenergic Receptors: Strosberg, A.D., Structure and function of the beta 3-adrenergic receptor. Annu Rev Pharmacol Toxicol, 1997. 37: p. 421-50.
pharmacology - what ligands there are and what their effects are
physiological and pathological roles