![]() Software Implementation
Software Implementation
Introduction
We implemented CHPM by Java/MATLAB cross-coding. The main interface is developed by Java and the core method is implemented by MATLAB. We design the software so that this cross platform implementation will be transparent to users as long as both Java Runtime Environment (JRE) and MATLAB are installed.
Features
Decompose gene expression time series data into biological processes defined by GO
Display a significant subset of biological processes and their corresponding activity levels for each dataset
Browse genes and their expression profiles assigned to each biological process
Browse biological processes and their activity levels associated with each gene
Display gene-process association table and their association weights
Automatically download and update the GO annotations
Learn the model from multiple datasets
System Requirements
Windows XP
JRE
MATLAB
Internet connection (for downloading the latest GO annotations)
How to set up
This software should be launched in MATLAB. Prior to using, please follow the following steps to set up.
Step 1. Download JRE and install.
Step 2. Download MATLAB and install.
Step 3. Set class path for MATLAB. Type 'edit classpath.txt' at the prompt. Add a new line for your local directory where you put the software.
![]()
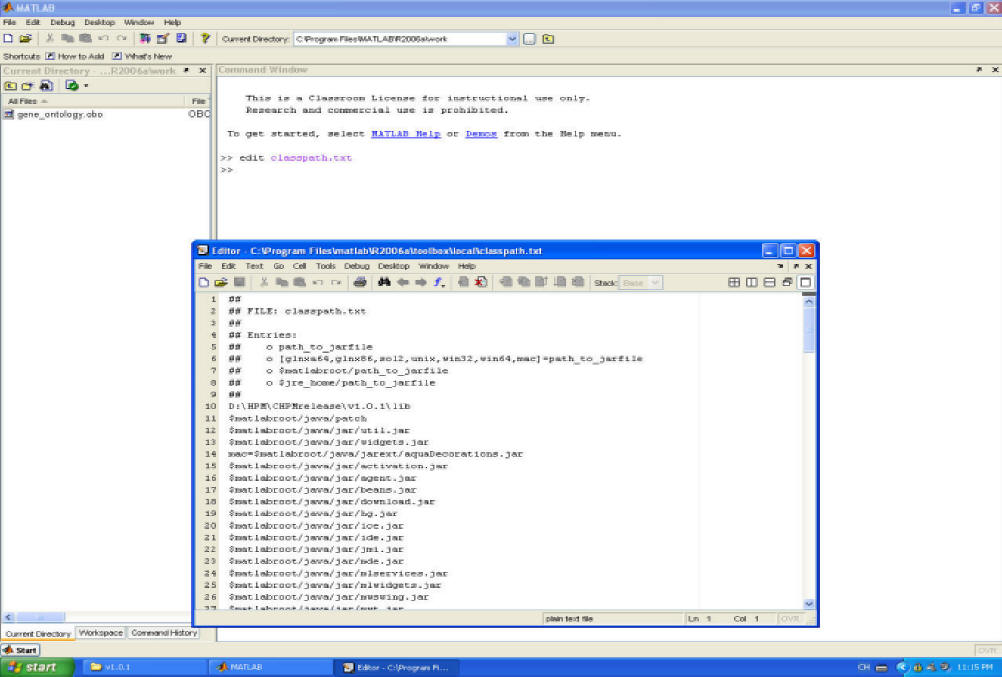
Step 4. Set path in MATLAB. Click 'file', next 'Set Path...', then add your local directory where you put the software to the list.

How to use
Step 1. Open MATLAB
Step 2. Simply enter 'runCHPM' at the prompt to run the software.
Step 3. Choose data file, gene annotation file, check 'Ontology' button (for the first time you use the software) for downloading the latest gene ontology structure file. Set parameters. Then, press 'Execute'.
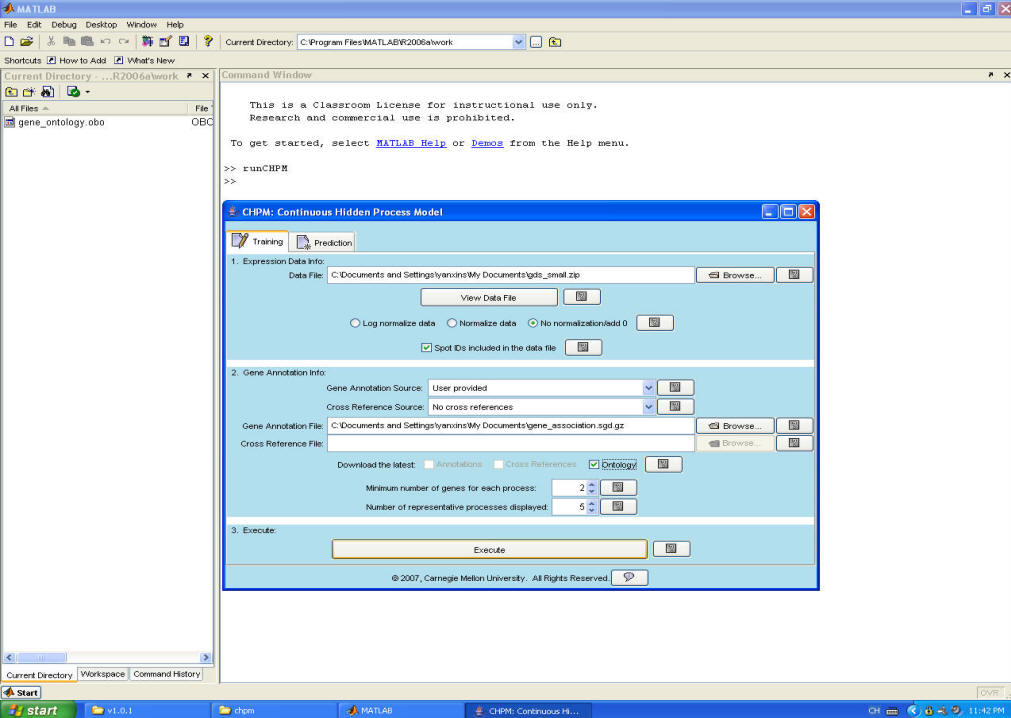
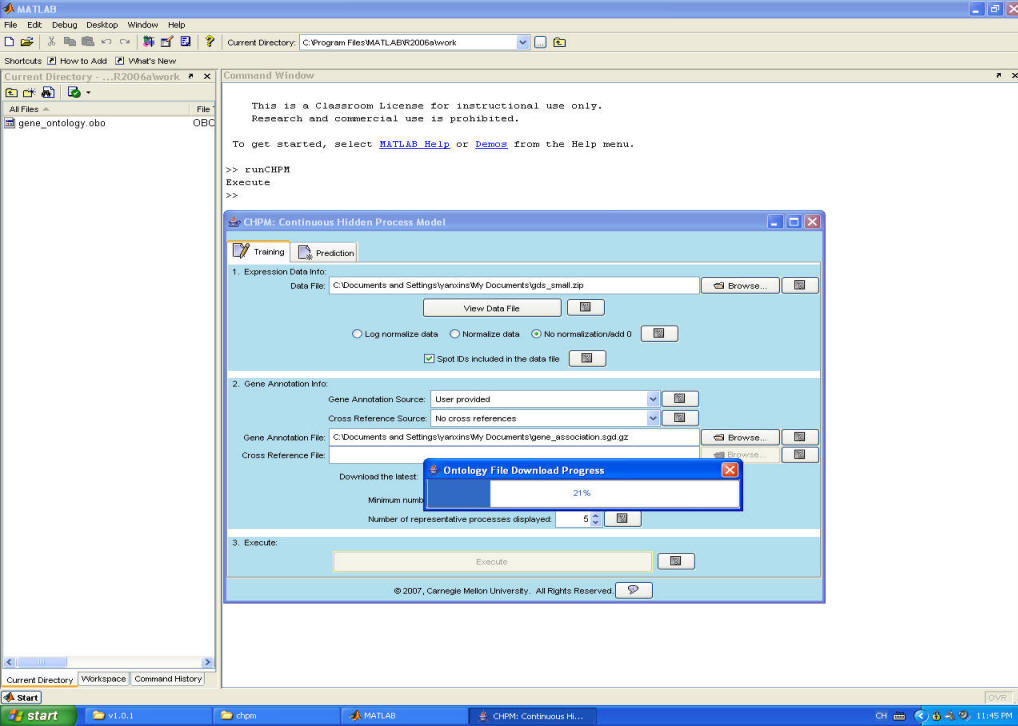
Step 4. The software will download the ontology file and use CHPM to infer the activity levels for hidden processes as well as the association between genes and processes.
Step 5. After computation, the results will be displayed. The main window displays the significant set of processes for each dataset.
Step 6. Press the yellow button besides each process to view the activity levels of this process and its associated genes expression levels.
Step 7. Press the yellow button "Display" to view the gene-process association tables.
Step 8. Click gene row to view the expression levels for the corresponding genes and the activity levels of associated processes in all datasets.
Releases
Version 1.1 --- released on 02/23/2007 [download] (added prediction part)
Version 1.0.2 --- released on 02/15/2007 [download] (fixed a bug in drawing the interface)
Version 1.0.1 --- released on 02/06/2007 [obsolete] (initial version)
Data
Here, we provide a small scale (but it's real) gene expression data which contains several hundreds yeast genes measured under 2 experimental conditions. We also provide a yeast gene GO annotations downloaded from SGD. For gene ontology structure data, users can check the 'ontology' option to let the software automatically download the latest version. We provide this data to assist users to familiarize themselves with the software. Users can download and play with this small scale data. Note that large scale data might takes quite a long time to analyze by CHPM.
Acknowledgement
The interface of this software adopts a significant part of source code from STEM.
The Matlab code was modified based on Kevin Murphy's BNT.