Kernel Methods for Large Scale Representation Learning
Additional Resources for Spectral Mixture Kernels and Fast
Kronecker Inference and Learning (new in GPML v3.5)
[Papers,
Lectures,
Code
and Tutorials]
Feel free to contact me (andrewgw@cs.cmu.edu) if you have any
questions.
This page underwent a major updates on
December 8, 2014, to include additional resources for the new
GPML v3.5,
NEW (July 4, 2015): Resources for Kronecker methods with
non-Gaussian likelihoods. Click
here
Spectral mixture kernels are motivated to help side-step kernel
selection. These kernels are especially useful on large
datasets. Structure
exploiting approaches, such as Kronecker methods, are particularly
suited to large scale kernel learning.
We are working on a major update to GPML, which will include KISS-GP/MSGP code. To match this release, we are also planning a release for deep kernel learning. We expect this code to be released by the end of June 2016.
Typo correction
regarding the spectral mixture kernel for multidimensional inputs
Andrew Gordon Wilson
July 4, 2015
Papers
Kernel Interpolation for Scalable Structured Gaussian Processes
(KISS-GP)
Andrew Gordon Wilson and Hannes Nickisch
To appear at the International Conference on Machine Learning (ICML),
2015
[PDF,
Supplement,
arXiv, BibTeX, Theme Song]
Fast kronecker inference in Gaussian processes with non-Gaussian
likelihoods
Seth Flaxman, Andrew Gordon Wilson, Daniel Neill, Hannes
Nickisch, and Alexander J. Smola
To appear at the International Conference on Machine
Learning (ICML), 2015
[PDF,
Supplement,
Code]
└ la carte - learning fast kernels
Zichao Yang, Alexander J. Smola, Le Song, and Andrew Gordon
Wilson
Artificial Intelligence and Statistics (AISTATS), 2015
Oral Presentation
[PDF, BibTeX]
Fast kernel learning for multidimensional pattern extrapolation
Andrew Gordon Wilson*, Elad Gilboa*, Arye Nehorai, and John P.
Cunningham
Advances in Neural Information Processing Systems (NIPS)
2014
[PDF, BibTeX, Code, Slides]
Covariance kernels for fast automatic pattern discovery and
extrapolation with Gaussian processes
Andrew Gordon Wilson
PhD Thesis, January 2014
[PDF,
BibTeX]
A process over all stationary covariance kernels
Andrew Gordon Wilson
Technical Report, University of Cambridge. June
2012.
[PDF]
Gaussian process kernels for pattern discovery and extrapolation
Andrew Gordon Wilson and Ryan Prescott Adams
International Conference on Machine Learning (ICML), 2013.
[arXiv, PDF,
Supplementary,
Slides,
Video
Lecture]
Lectures
- Abstract,
Slides, and Audio, for the lecture "Kernel Methods for Large
Scale Representation Learning", on
November 5, 2014, at Oxford University. This lecture covers
material on spectral mixture kernels and fast
Kronecker methods as part of a high level vision for building kernel
methods for large scale representation learning.
- An introductory tutorial lecture on the spectral mixture kernel
can be found here.
This lecture was presented on
June 2013 at ICML in Atlanta, Georgia.
Code and Tutorials
The GPML
package includes Matlab and Octave compatible code for fast
grid methods and spectral mixture kernels.
All tutorial files can be downloaded
together as part of this zip repository.
Spectral Mixture Kernels:
- covGabor and covSM (updated in GPML v3.5, December 8, 2014)
- covSMfast.m (a fast implementation)
- spectral_init.m: an initialisation script for the spectral mixture
kernel (can improve robustness of results to a wide range of
problems)
Fast Kronecker Inference
- covGrid and infGrid (introduced in GPML v3.5, December 8, 2014)
Fast Kronecker Inference with non-Gaussian Likelihoods
- infGrid_Laplace.m
(introduced July 4, 2015)
Tutorial Files
- Image Extrapolation and Kernel Learning
(vincent.m)
- covGrid and infGrid accuracy test (test_covGrid.m,
test_infGrid.m, courtesy of Hannes Nickisch)
- Time Series Extrapolation Experiment
- Datasets: Treadplate, Wood, and NIPS patterns, CO2 and Airline
Time Series, Hickories
Structure Discovery Tutorial
Download files and add to path
1) Download
GPML v3.5 and add the files to the Matlab (or Octave
path).
2) For the tutorials, the only required external files are
'vincent.m', 'spectral_init.m',
'test_covGrid.m', 'test_infGrid.m', and the datasets.
'covSMfast.m' is an optional faster
implementation of covSM. All
tutorial files are available in this zip file.
Unzip these
tutorial files into one directory.
Example 1: Try running 'test_covGrid.m' and
'test_infGrid.m' . These show the accuracy
of the grid methods compared to standard methods involving
Cholesky decompositions.
covGrid is used to generate a covariance matrix with Kronecker
product structure, which
can be exploited by infGrid. This test shows the
generated covariance matrix is identical
to what would be obtained using standard methods.
infGrid exploits Kronecker structure for scalable inference and
learning. In the case where
the inputs are on a grid, with no missing inputs, the inference
and learning is also exact. In
this demo, the data only has partial grid
structure. Every second data point is missing, as
indicated by idx = (1:2:N)'. The idx
variable contains the locations of the training points.
We use the extensions in Wilson
et. al (2014) to handle partial grids
efficiently. With partial
grids, inference requires linear conjugate gradients
(LCG). With a high tolerance (< 1e-4),
and number of iterations (> 400), inference is exact to
within numerical precision. Typically,
a tolerance of 1e-2 and maximum iterations of 200 is sufficient.
With partial grids, the log
determinant of the covariance matrix, used for marginal
likelihood evaluations, undergoes
a small approximation.
Exercise 1: Try adjusting opt.cg_maxit and opt.cg_tol, and
observe the effect on inference.
Exercise 2: Try changing idx
to idx = (1:1:N)' and see what happens.
Exercise 3: Cycle between the three test cases, cs = 1,
2, 3.
Example 2: Try running 'vincent.m'. You will be
given a choice of patterns, and the code
will extrapolate these patterns automatically. This script
reconstructs some of the results in
Fast
Kernel Learning for Multidimensional Pattern Extrapolation.
Do not worry if there are occasional warnings that LCG has
not converged. The
optimization will proceed normally. The warnings can be
resolved by adjusting
the tolerance and maximum number of iterations. We have
set these values for a
good balance of accuracy and efficiency, not minding if LCG
occasionally does
not converge to within the specified precision.
Some example output:

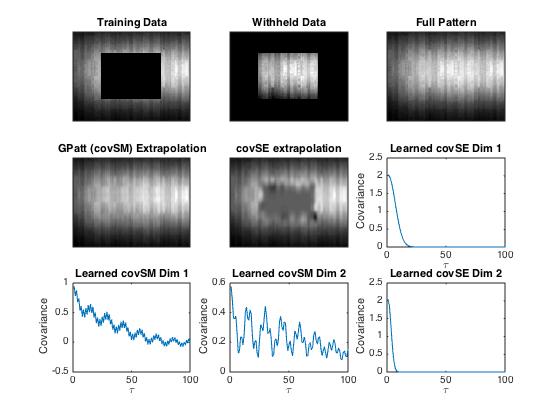
Note the learned spectral mixture kernels. It would be
difficult to know to a priori hard code
kernels like these. The learned structure is interpretable
and enables good predictions and
extrapolation.
Exercise 1: Extrapolations are produced with spectral
mixture kernels and SE kernels.
Try adding a Matern kernel to this demo. It will do
better on the letters example than
an SE kernel, but not as well as the spectral mixture kernel
on larger missing regions.
Exercise 2: Study the initialisation script
spectral_init.m. This is a basic and
robust initialisation for spectral mixture kernels. Can
you think of other initialisations
which might offer even better performance?
Exercise 3: Reproduce Figure 1j in Wilson et. al,
NIPS 2014, showing the initial
and learned weights against the initial and learned
frequencies.
Exercise 4: Adjust this script to your own patterns and
general purpose regression
problems, such as the land surface temperature forecasting in
Wilson et. al, NIPS 2014.
Example 3: Time Series Extrapolation
Files are forthcoming.
We provide CO2 and airline time series datasets, with representative
results shown below. Training data are represented in blue,
testing
data in green, and extrapolations using GPs with SM and SE (squared
exponential) kernels are in black and red, respectively. The
gray shade represents the
95% posterior credible interval using a Gaussian process with an SM
kernel.


Non-Gaussian Likelihood Tutorial
This tutorial is
based on the paper:
Fast kronecker inference in Gaussian processes with
non-Gaussian likelihoods
Seth Flaxman, Andrew Gordon Wilson, Daniel
Neill, Hannes Nickisch, and Alexander J. Smola
To appear at the International Conference on
Machine Learning (ICML), 2015
[PDF,
Supplement]
Files
Download infGrid_Laplace.m
Contents
Hickory data
Author: Seth Flaxman 4 March
2015
clear all, close all
xy = csvread('hickories.csv');
plot(xy(:,1),xy(:,2),'.')
n1 = 32; n2 = 32;
x1 = linspace(0,1.0001,n1+1); x2 = linspace(-0.0001,1,n2+1);
xg = {(x1(1:n1)+x1(2:n1+1))'/2, (x2(1:n2)+x2(2:n2+1))'/2};
i = ceil((xy(:,1)-x1(1))/(x1(2)-x1(1)));
j = ceil((xy(:,2)-x2(1))/(x2(2)-x2(1)));
counts = full(sparse(i,j,1));
cv = {@covProd,{{@covMask,{1,@covSEiso}},{@covMask,{2,@covSEisoU}}}};
cvGrid = {@covGrid, { @covSEiso, @covSEisoU}, xg};
hyp0.cov = log([.1 1 .1]);
y = counts(:);
X = covGrid('expand', cvGrid{3});
Idx = (1:length(y))';
lik = {@likPoisson, 'exp'};
hyp0.mean = .5;

Laplace approximation without grid structure
tic
hyp1 = minimize(hyp0, @gp, -100, @infLaplace, @meanConst, cv, lik, X, y);
toc
exp(hyp1.cov)
Function evaluation 0; Value 1.114380e+03
Function evaluation 5; Value 1.104834e+03
Function evaluation 8; Value 1.098415e+03
Function evaluation 12; Value 1.097589e+03
Function evaluation 16; Value 1.097445e+03
Function evaluation 18; Value 1.097417e+03
Function evaluation 23; Value 1.096718e+03
Function evaluation 26; Value 1.096703e+03
Function evaluation 30; Value 1.096424e+03
Function evaluation 33; Value 1.096419e+03
Function evaluation 35; Value 1.096419e+03
Function evaluation 37; Value 1.096419e+03
Function evaluation 46; Value 1.096419e+03
Elapsed time is 300.589011 seconds.
ans =
0.0815 0.6522 0.0878
Laplace approximation with grid structure and Kronecker
inference
tic
hyp2 = minimize(hyp0, @gp, -100, @infGrid_Laplace, @meanConst, cvGrid, lik, Idx, y);
toc
exp(hyp2.cov)
Function evaluation 0; Value 1.151583e+03
Function evaluation 12; Value 1.124633e+03
Function evaluation 14; Value 1.116309e+03
Function evaluation 17; Value 1.115676e+03
Function evaluation 20; Value 1.115373e+03
Function evaluation 23; Value 1.115308e+03
Elapsed time is 41.778548 seconds.
ans =
0.2346 0.5824 0.1100
Model comparison
The models learned slightly different hyperparameters. Are the
likelihoods different?
fprintf('Log-likelihood learned w/out Kronecker: %.04f\n', gp(hyp1, @infLaplace, @meanConst, cv, lik, X, y));
fprintf('Log-likelihood learned with Kronecker: %.04f\n', gp(hyp2, @infLaplace, @meanConst, cv, lik, X, y));
fprintf('Log-likelihood with Kronecker + Fiedler bound: %.04f\n', gp(hyp2, @infGrid_Laplace, @meanConst, cvGrid, lik, Idx, y));
[Ef1 Varf1 fmu1 fs1 ll1] = gp(hyp1, @infGrid_Laplace, @meanConst, cvGrid, lik, Idx, y, Idx, y);
[Ef2 Varf2 fmu2 fs2 ll2] = gp(hyp2, @infGrid_Laplace, @meanConst, cvGrid, lik, Idx, y, Idx, y);
figure(1); imagesc([0 1], [0 1], (reshape(Ef1,32,32)')); set(gca,'YDir','normal'); colorbar;
figure(2); imagesc([0 1], [0 1], (reshape(Ef2,32,32)')); set(gca,'YDir','normal'); colorbar;
Log-likelihood learned w/out Kronecker: 1096.4188
Log-likelihood learned with Kronecker: 1100.3819
Log-likelihood with Kronecker + Fiedler bound: 1115.3084


Kronecker + grid enables a finer resolution
now let's exploit the grid structure to get a finer resolution
map we'll also switch covariances, just for fun.
hyp.cov = log([.3 1 .2 1]);
lik = {@likPoisson, 'exp'}; hyp.lik = [];
mean = {@meanConst}; hyp.mean = .5;
n1 = 64; n2 = 64;
x1 = linspace(0,1.0001,n1+1); x2 = linspace(-0.0001,1,n2+1);
xg = {(x1(1:n1)+x1(2:n1+1))'/2, (x2(1:n2)+x2(2:n2+1))'/2};
i = ceil((xy(:,1)-x1(1))/(x1(2)-x1(1)));
j = ceil((xy(:,2)-x2(1))/(x2(2)-x2(1)));
counts = full(sparse(i,j,1));
y = counts(:);
xi = (1:numel(y))';
cov = {@covGrid, { {@covMaterniso,5}, {@covMaterniso,5}}, xg};
opt.cg_maxit = 400; opt.cg_tol = 1e-2;
inf_method = @(varargin) infGrid_Laplace(varargin{:},opt);
tic
hyp2 = minimize(hyp, @gp, -100, inf_method, mean, cov, lik, xi, y(:));
toc
exp(hyp2.cov)
[post nlZ dnlZ] = infGrid_Laplace(hyp2, mean, cov, lik, xi, y(:));
post.L = @(x) 0*x;
ymu = gp(hyp2, @infGrid_Laplace, mean, cov, lik, xi, post, xi);
figure(3), imagesc([0 1], [0 1], reshape(ymu,size(counts))'),
hold on;
set(gca,'YDir','normal');
plot(xy(:,1),xy(:,2),'w.');
colorbar
hold off
Function evaluation 0; Value 1.954834e+03
Function evaluation 6; Value 1.946841e+03
Function evaluation 8; Value 1.940877e+03
Function evaluation 11; Value 1.940012e+03
Function evaluation 14; Value 1.938218e+03
Function evaluation 17; Value 1.936884e+03
Function evaluation 20; Value 1.934900e+03
Function evaluation 22; Value 1.933195e+03
Elapsed time is 187.843026 seconds.
ans =
0.3051 0.7512 0.1636 0.7512

Published with
MATLAB« R2013a
Acknowledgements
Thanks to Hannes Nickisch, Seth Flaxman, Carl Edward
Rasmussen, Elad Gilboa, and John P. Cunningham for
their feedback
and assistance. Thanks to Hannes in particular for many of the
new features in GPML v3.5.







